
To create the network we should state Source Interaction and Target Interaction. Open Cytoscape and import the downloaded file into the tool by Import > Network > File.
#Network analyzer cytoscape download#
To download the data, follow Tables/Exports > simple tabular text output. Expand the network by following the steps, Data settings > max number of interactors > custom value > max interactors = 60. You’ll see a small network of 10 interacting proteins. Search for Protein name: "Alzheimer" and set the organism: “homo sapiens”. Now we have a file that we can investigate! We need to select at least two parameters protein name/id and chemical name/id.Ĭreate the report and download it one of the given formats. Since we want to build a protein-ligand interaction network, our source/target node is protein whereas target/source node is chemical.
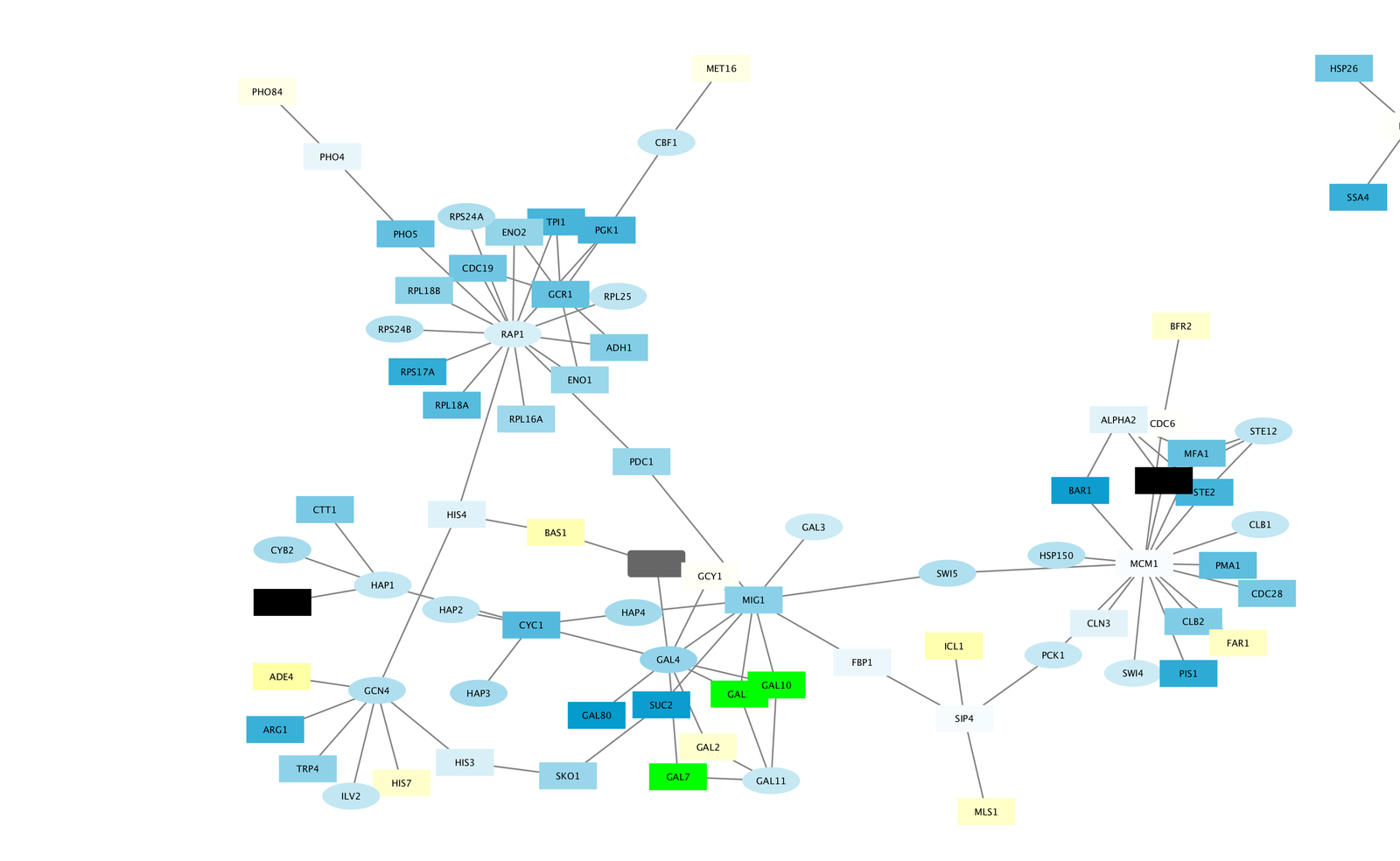
To be able to define an interaction we need source and target nodes. Go to Reports menu and select Custom Reports > Customizable Table.Ĭhoose the parameters you are interested in. You might filter the results by selecting a specific organism. Set the second parameter as HasLigand(s) then select Yesīy this way, we make sure that each protein in this data set binds to at least one ligand. Set the first parameter as Text, then type Diabetes Mellitus. In the closeness centrality, the node that spread information to the others as fast as possible is the most central one in this case where the fast defined as short paths.Ĭlustering is the task of dividing a set of objects into groups/clusters/cliques so that the objects with similar properties are placed together.Ĭlustering helps us to identify densely connected nodes.Įxample Tasks Protein-Ligand Interaction Data Degreeĭegree centrality is simply measured by the number of ties that a node has.īetweenness centrality quantifies the number of times a node acts as a bridge along the shortest Importance decreases in the following direction: bigger, red -> smaller, green. In the examples below, size and color of the nodes indicate importance. CentralityĬentrality metrics decide how important an actor in the network based on its position. Data set contains total 34 actors (nodes) with 78 interactions (edges). Zachary Karate Club, a well-known social network data set of university karate club, is used for demonstration. GLASS is a GPCR-ligand interaction database.STITCH provides three different interaction types to invesgate, chemical-chemical, protein-chemical and protein-protein.
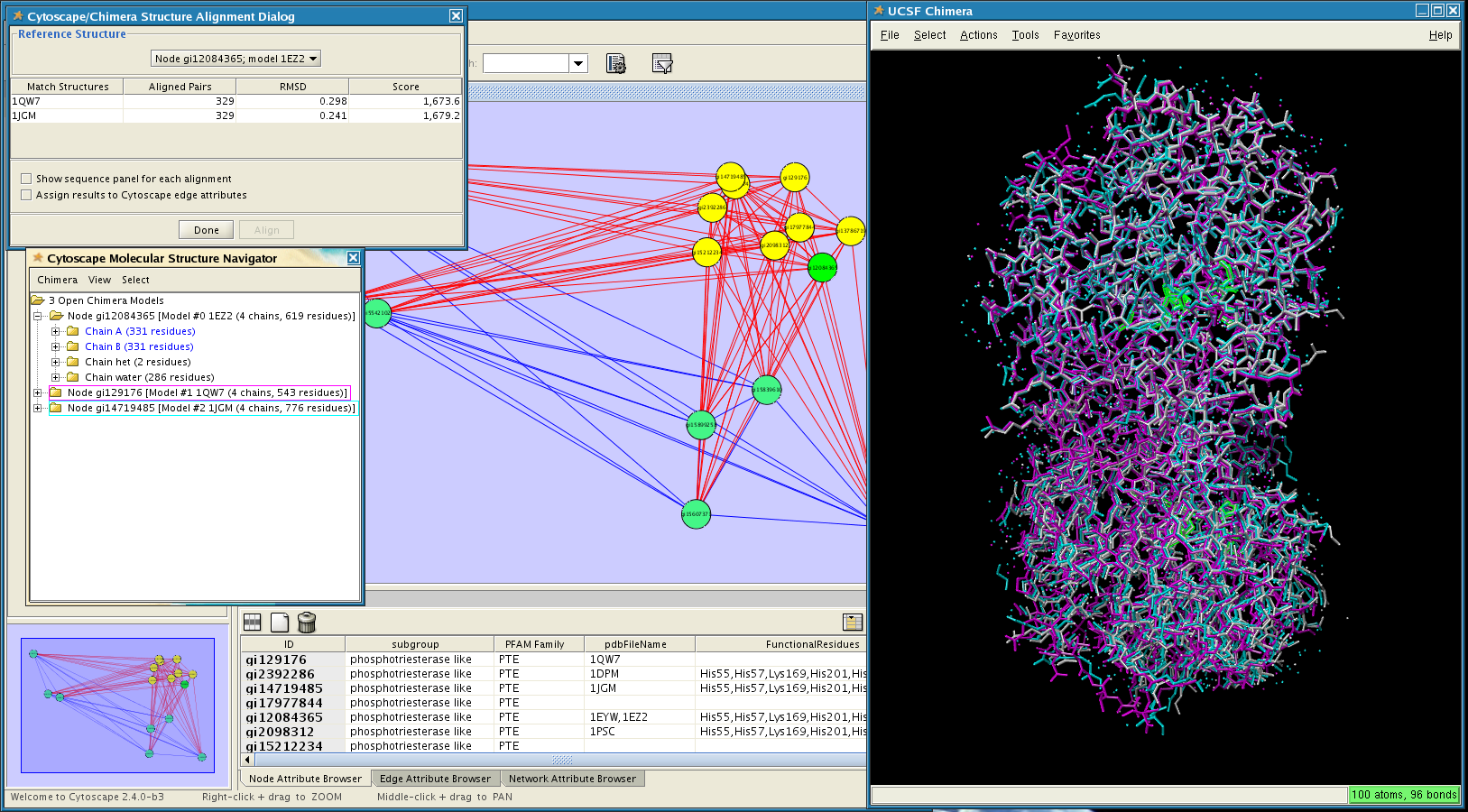
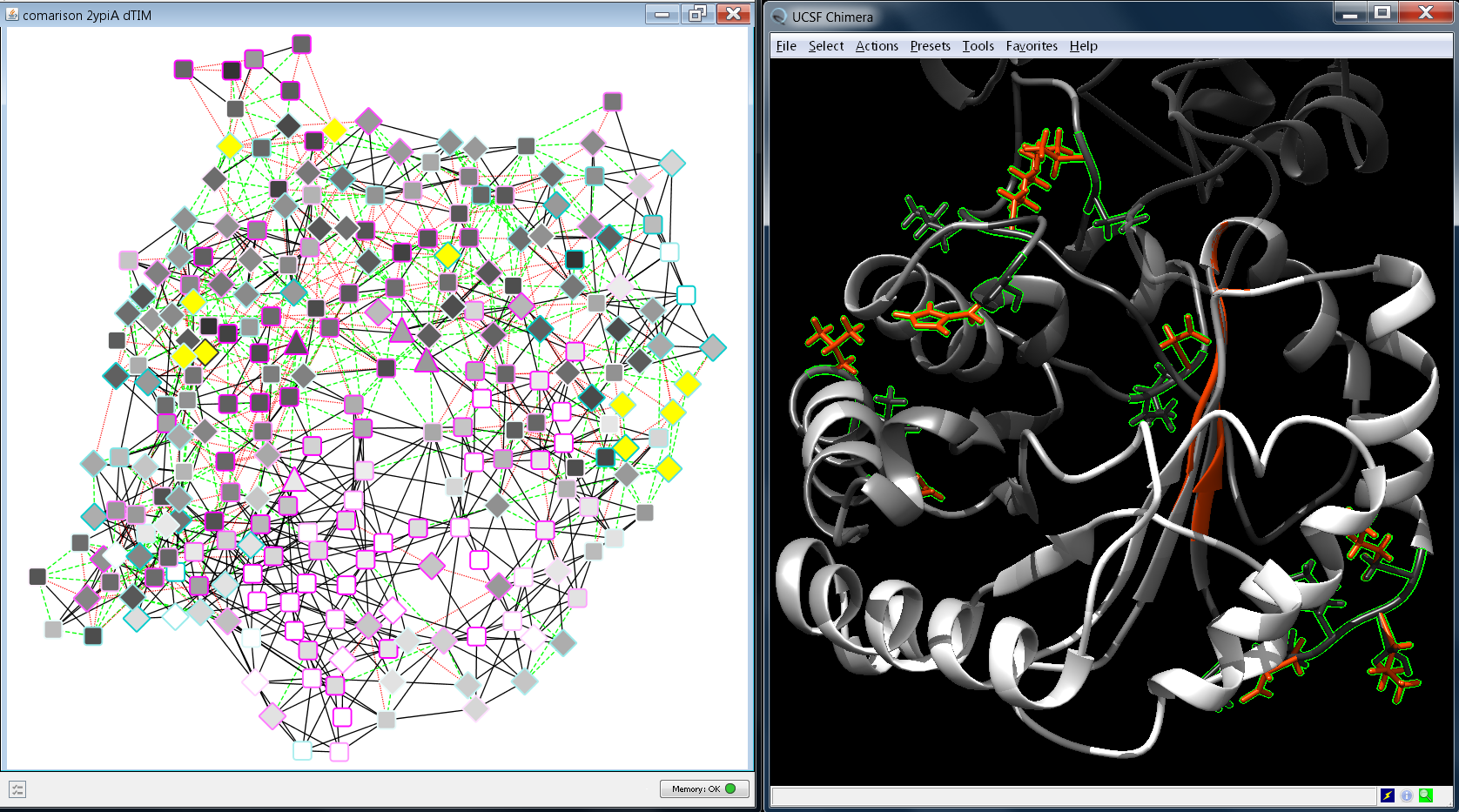
MCODE is one of the specific methods to identify clusters.ClusterMaker2 provides a wide range of algorithms to find the densely connected clusters for the given network.Network Analyzer is a built-in plugin of Cytoscape, which provides statistical parameters of the network as well as central nodes.Cytoscape enables use of broad range of plugins targeting clustering, graph analysis,.Cytoscape is an open source environment for the analysis and.

Some other useful tutorials, Analysis and Visualization of BiologicalNetworks with Cytoscape and Introduction to Cytoscape 3.1. Cytoscape version 3.2.1 was used in this tutorial. This tutorial aims to help to beginners in the Bioinformatics fields to use Cytoscape and utilize some important network parameters such as centrality and clustering.


 0 kommentar(er)
0 kommentar(er)
